[R package] Embedding Key Descriptive Statistics within Original Data (Feat. descriptivestat)
When analyzing data, we often need to examine descriptive statistics such as standard deviation, standard error, or the coefficient of variation (CV). These are typically summarized in a short table, but in some cases, it may be necessary to include such statistics directly in the original dataset for further analysis.
Let’s upload a dataset to begin.
if(!require(readr)) install.packages("readr")
library(readr)
github="https://raw.githubusercontent.com/agronomy4future/raw_data_practice/refs/heads/main/4_treatments_4_genotypes_with_4_blocks.csv"
df=data.frame(read_csv(url(github), show_col_types=FALSE))
head(df,5)
Genotype Block Treatment Yield
1 CV1 I I 42.9
2 CV1 II I 41.6
3 CV1 III I 28.9
4 CV1 IV I 30.8
5 CV2 I I 53.3
.
.
.I would like to calculate the mean, variance, standard deviation, standard error, 95% confidence interval, coefficient of variation, and the 25th percentile of yield data, grouped by genotype and treatment. I will use the following R code to perform these calculations.
if(!require(dplyr)) install.packages("dplyr")
library(dplyr)
summary_stats= df %>%
group_by(Genotype, Treatment) %>%
summarise(
n= length(Yield),
mean= mean(Yield, na.rm= TRUE),
variance= var(Yield, na.rm= TRUE),
sd= sd(Yield, na.rm= TRUE),
se= sd / sqrt(n),
ci_lower= mean - qt(0.975, df= n-1)*se,
ci_upper= mean + qt(0.975, df= n-1)*se,
cv= sd/mean,
q25= quantile(Yield, probs= 0.25, na.rm=TRUE),
.groups= "drop")
print(summary_stats)
Genotype Treatment n mean variance sd se ci_lower ci_upper cv q25
1 CV1 I 4 36.0 52.1 7.22 3.61 24.6 47.5 0.200 30.3
2 CV1 II 4 50.6 45.3 6.73 3.37 39.9 61.3 0.133 45.7
3 CV1 III 4 45.8 48.2 6.94 3.47 34.8 56.9 0.151 40.4
4 CV1 IV 4 37.3 52.8 7.27 3.63 25.7 48.9 0.195 33.1
5 CV2 I 4 50.8 212. 14.6 7.28 27.7 74.0 0.286 42.8
6 CV2 II 4 55.4 129. 11.4 5.68 37.3 73.5 0.205 49.5
7 CV2 III 4 53.1 134. 11.6 5.79 34.7 71.5 0.218 44.4
8 CV2 IV 4 54.3 72.3 8.50 4.25 40.8 67.8 0.157 49.7
9 CV3 I 4 53.9 63.7 7.98 3.99 41.2 66.6 0.148 48.9
10 CV3 II 4 51.4 69.3 8.33 4.16 38.1 64.6 0.162 46.3
11 CV3 III 4 55.9 82.1 9.06 4.53 41.5 70.3 0.162 49.2
12 CV3 IV 4 56.0 28.8 5.36 2.68 47.5 64.6 0.0957 52.5
13 CV4 I 4 61.9 114. 10.7 5.35 44.9 78.9 0.173 53.7
14 CV4 II 4 63.4 40.2 6.34 3.17 53.3 73.5 0.0999 58.3
15 CV4 III 4 57.7 124. 11.1 5.57 39.9 75.4 0.193 49.6
16 CV4 IV 4 61.2 129. 11.4 5.68 43.2 79.3 0.185 54.3However, I would like to append these descriptive statistics to my raw dataset (df) in order to include them in my figures. To facilitate this process, I created a new R package called descriptivestat(). Below is the basic structure of the code:
| data | A data frame containing the input dataset. |
| group_vars | Character vector of column names used to group the data |
| value_vars | Character vector of numeric variable names to compute statistics |
| output_stats | Character vector of statistics to compute. Valid options include: mean is automatically calculated and inserted to the rows v: Variance sd: Standard deviation se: Standard error ci: 95% confidence interval (LCI, UCI) cv: Coefficient of variation iqr: Interquartile range (Q1, Q2, Q3) |
# upload required package
if(!require(remotes)) install.packages("remotes")
if (!requireNamespace("descriptivestat", quietly = TRUE)) {
remotes::install_github("agronomy4future/descriptivestat", force= TRUE)
}
library(remotes)
library(descriptivestat)I will calculate the mean, standard deviation, coefficient of variation, interquartile range, and other descriptive statistics, and append them to the original dataset.
# to calculate descriptive statistics
df1= descriptivestat(data= df, group_vars= c("Genotype", "Treatment"),
value_vars= c("Yield"),
output_stats= c("v","sd","se","ci","cv","iqr"))
print(df1, n= Inf)This package inserts the mean values between data rows by group, and the corresponding descriptive statistics are also added to the raw dataset.
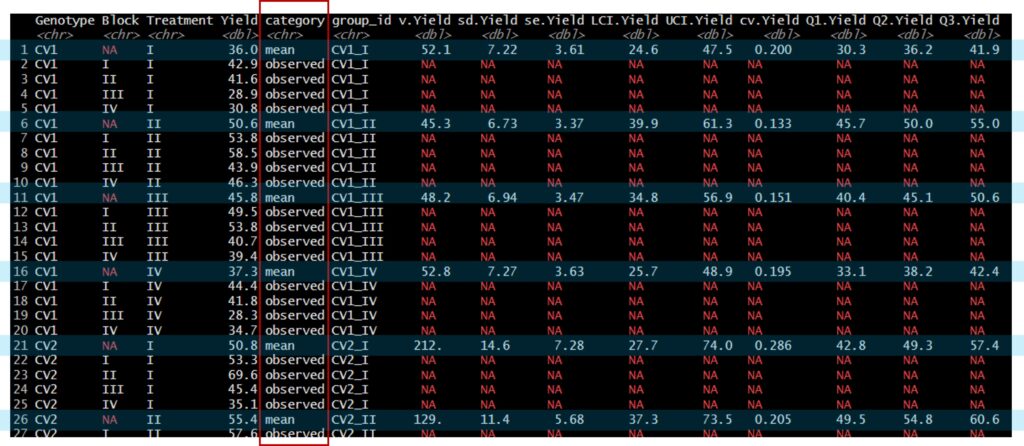
Github: https://github.com/agronomy4future/descriptivestat
I’ll create a figure.
Fig1= ggplot() +
geom_errorbar(data= subset(df1, category=="mean"),
aes(x= Genotype, ymin= Yield-se.Yield, ymax=Yield+se.Yield),
width= 0.1, color= "black") +
geom_point(data= subset(df1, category=="mean"),
aes(x= Genotype, y= Yield, fill= Treatment, shape=Treatment),
size=4, color="black") +
scale_fill_manual(name="Treatment", values= c("I"="red", "II"="purple", "III"="orange", "IV"="blue")) +
scale_shape_manual(name="Treatment", values = c("I"=21, "II"=22, "III"=23, "IV"=24)) +
geom_jitter(data= subset(df1, category=="observed"),
aes(x= Genotype, y= Yield), width=0.1, alpha=0.6,
size=2, color="gray40") +
scale_y_continuous(breaks=seq(0,100,20), limits = c(0,100)) +
labs(x= NULL, y="Yield") +
theme_classic(base_size=18, base_family="serif") +
theme(
legend.position="right",
legend.key=element_rect(color="white", fill="white"),
legend.text=element_text(family="serif", face="plain",
size=15, color= "black"),
legend.background=element_rect(fill=alpha("white", 0.5)),
strip.background= element_rect(color= "black", fill="grey75",
linewidth= 0.5, linetype="solid"),
panel.border= element_rect(color="black", fill=NA, linewidth=0.5),
panel.grid.major= element_line(color="grey90", linetype="dashed"),
axis.line= element_blank())
options(repr.plot.width=9, repr.plot.height=5)
print(Fig1)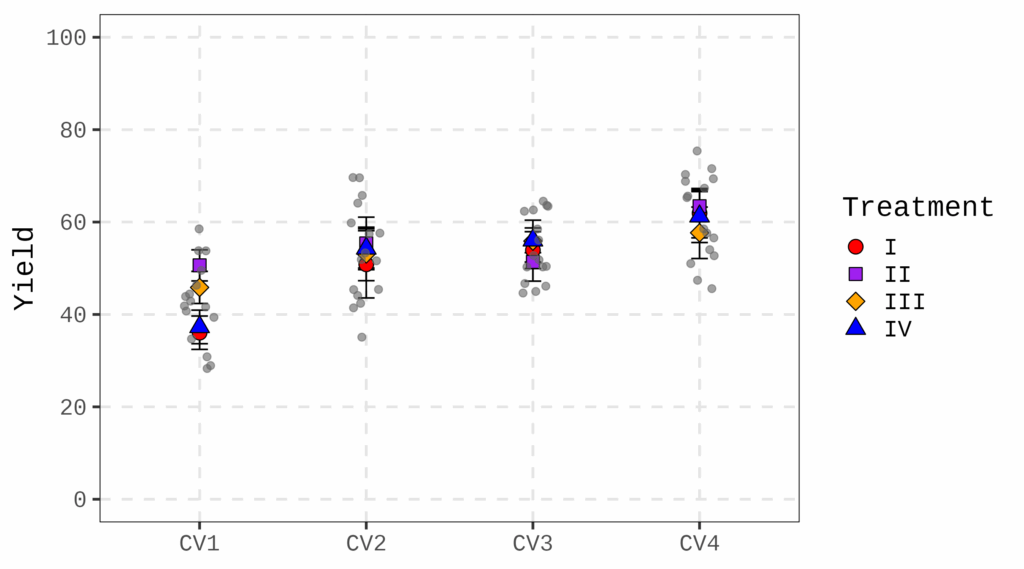
If you would like to use standard deviation as the error bar, you can simply modify the code accordingly.
geom_errorbar(data= subset(df1, category=="mean"),
aes(x= Genotype, ymin= Yield-sd.Yield, ymax=Yield+sd.Yield),
width= 0.1, color= "black") +This dataset enables visualization of both the raw data points and their means, along with standard deviation, standard error, or other descriptive statistics.

Here is another example using a large dataset.
if(!require(readr)) install.packages("readr")
library(readr)
github="https://raw.githubusercontent.com/agronomy4future/raw_data_practice/refs/heads/main/wheat_grain_grea_and_heat_tolerance.csv"
df=data.frame(read_csv(url(github), show_col_types=FALSE))
df = subset(df, select = -tolerance)
head(df,5)
genotype thinning area
1 cv1 control 7.56
2 cv1 control 7.25
3 cv1 control 7.99
4 cv1 control 5.86
5 cv1 control 8.04
.
.
.
tail(df,5)
genotype thinning area
7970 cv2 manipulation 19.71
7971 cv2 manipulation 16.40
7972 cv2 manipulation 21.38
7973 cv2 manipulation 14.33
7974 cv2 manipulation 10.83 This is a large dataset used to measure wheat grain area in response to a thinning treatment applied in the field. In each plot, alternate wheat rows were removed to increase assimilate availability by reducing competition. Due to the large number of data points (7,974 datapoints), visualizing individual raw data is not practical; instead, boxplots or bar graphs would be more appropriate.
Fig2= ggplot(data=df, aes(x= genotype, y=area)) +
geom_boxplot(aes(fill= thinning, color= thinning), color="black") +
scale_fill_manual(name="Thinning", values= c("control"="orange", "manipulation"="darkred")) +
scale_y_continuous(breaks=seq(0,30,10), limits= c(0,30)) +
facet_wrap(~ thinning, scales= "free") +
labs(x= NULL, y="Wheat grain area") +
theme_classic(base_size=18, base_family="serif") +
theme(
legend.position="none",
legend.key=element_rect(color="white", fill="white"),
legend.text=element_text(family="serif", face="plain",
size=15, color= "black"),
legend.background=element_rect(fill=alpha("white", 0.5)),
strip.background= element_rect(color= "black", fill="grey75",
linewidth= 0.5, linetype="solid"),
panel.border= element_rect(color="black", fill=NA, linewidth=0.5),
panel.grid.major= element_line(color="grey90", linetype="dashed"),
axis.line= element_blank())
options(repr.plot.width=9, repr.plot.height=5)
print(Fig2)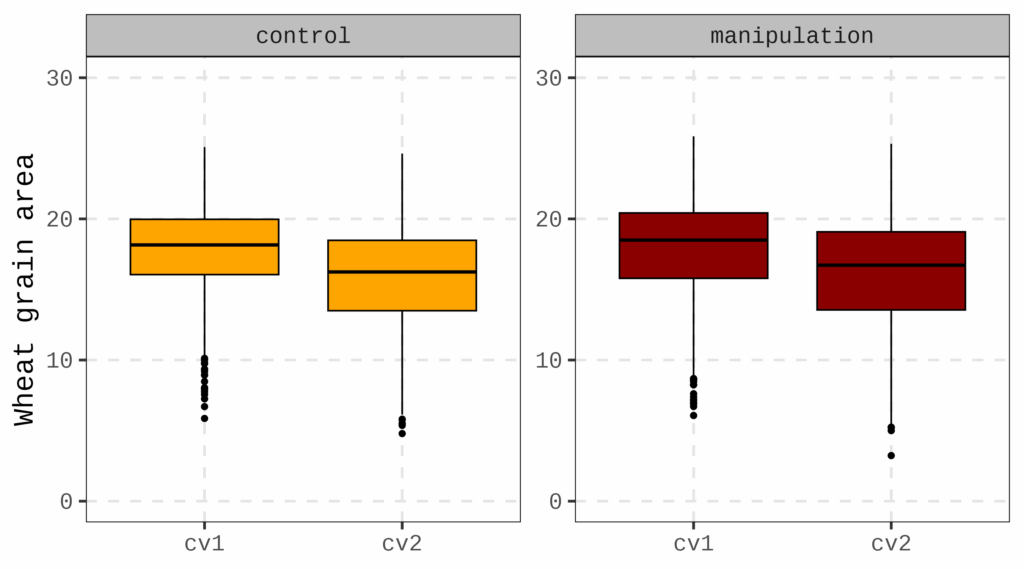

However, I would like to visualize both the raw data and its variability. In this case, the descriptivestat() package is useful for that purpose.
First, I will output the standard deviation and interquartile range (IQR) for this dataset.
df1= descriptivestat(data= df, group_vars= c("genotype", "thinning"),
value_vars= c("area"),
output_stats= c("sd","iqr"))
print(head(df1, 5))
genotype thinning area category group_id sd.area Q1.area Q2.area Q3.area
1 cv1 control 17.8 mean cv1_control 2.97 16.1 18.2 20.0
2 cv1 control 7.56 observed cv1_control NA NA NA NA
3 cv1 control 7.25 observed cv1_control NA NA NA NA
4 cv1 control 7.99 observed cv1_control NA NA NA NA
5 cv1 control 5.86 observed cv1_control NA NA NA NA
.
.
. This graph will help illustrate both individual variability and the overall trend in the data.
Fig3= ggplot() +
# Observed data (gray jittered points)
geom_jitter(data= subset(df4, category=="observed"),
aes(x= genotype, y= area, fill=genotype, shape=thinning), width=0.2,
alpha=0.7, size=2, color="gray75") +
# Error bars for means
geom_errorbar(data= subset(df4, category=="mean"),
aes(x= genotype, ymin= area-sd.area, ymax=area+sd.area),
width= 0.1, color= "black") +
# Mean points (colored)
geom_point(data= subset(df4, category=="mean"),
aes(x= genotype, y= area, fill=genotype, shape=thinning),
size=4, color="black") +
scale_fill_manual(values= c("cadetblue", "darkred")) +
scale_shape_manual(name="Thinning", values = c("control"=21, "manipulation"=22)) +
scale_y_continuous(breaks=seq(0,30,10), limits = c(0,30)) +
facet_wrap(~ thinning, scales = "free") +
coord_flip () +
labs(x= NULL, y="Wheat grain area") +
theme_classic(base_size=18, base_family="serif") +
theme(
legend.position="none",
#legend.title=element_blank(),
#axis.text.x = element_text(angle= 90, vjust= 0.5, hjust= 1),
legend.key=element_rect(color="white", fill="white"),
legend.text=element_text(family="serif", face="plain",
size=15, color= "black"),
legend.background=element_rect(fill=alpha("white", 0.5)),
strip.background= element_rect(color= "black", fill="grey75",
linewidth= 0.5, linetype="solid"),
panel.border= element_rect(color="black", fill=NA, linewidth=0.5),
panel.grid.major= element_line(color="grey90", linetype="dashed"),
axis.line= element_blank()
)
options(repr.plot.width=9, repr.plot.height=5)
print(Fig3)
ggsave("Fig3.png", plot= Fig3, width=9, height= 5, dpi= 300)

Since the descriptivestat() package appends the mean and other descriptive statistics to the raw dataset, we can easily use them for visualization.
If you want to use upper and lower percentiles as error bars, you can simply modify the code as shown below.
geom_errorbar(data= subset(df1, category=="mean"),
aes(x= genotype, ymin= area-(area-Q1.area), ymax=area+(Q3.area-area)),
width= 0.1, color= "black") +
I’ll create boxplot using descriptivestat() package.
Fig4= ggplot(data=df3, aes(x= genotype, y=area)) +
geom_jitter(aes(x= genotype, y= area, fill=genotype, shape=thinning),
width=0.2, alpha=0.7, size=2, color="gray75") +
geom_boxplot(aes(fill= genotype, color= thinning), color="black") +
scale_fill_manual(values= c("cadetblue", "darkred")) +
scale_shape_manual(values= c(21,22)) +
scale_y_continuous(breaks=seq(0,30,10), limits = c(0,30)) +
coord_flip () +
facet_wrap(~ thinning, scales = "free") +
labs(x= NULL, y="Wheat grain area (mm2)") +
theme_classic(base_size=18, base_family="serif") +
theme(
legend.position="none",
legend.key=element_rect(color="white", fill="white"),
legend.text=element_text(family="serif", face="plain",
size=15, color= "black"),
legend.background=element_rect(fill=alpha("white", 0.5)),
strip.background= element_rect(color= "black", fill="grey75",
linewidth= 0.5, linetype="solid"),
panel.border= element_rect(color="black", fill=NA, linewidth=0.5),
panel.grid.major= element_line(color="grey90", linetype="dashed"),
axis.line= element_blank())
options(repr.plot.width=9, repr.plot.height=5)
print(Fig4)
ggsave("Fig4.png", plot= Fig4, width=9, height= 5, dpi= 300)
When compared to a boxplot, the upper and lower percentiles are exactly the same as those represented by the box boundaries.

Which type of graph do you prefer? While a boxplot effectively summarizes data variability using quartiles, the graph generated with the descriptivestat() package provides a clearer visualization by displaying the actual raw data alongside the mean and other descriptive statistics.
full code: r_code/Embedding_Key_Descriptive_Statistics_within_Original_Data_(Feat__descriptivestat).ipynb at main · agronomy4future/r_code

We aim to develop open-source code for agronomy ([email protected])
© 2022 – 2025 https://agronomy4future.com – All Rights Reserved.
Last Updated: 18/05/2025