[R package] An easy way to use interpolation code to predict in-between data points
In my previous post, I explained how to calculate interpolation to predict in-between data points.
■ [Data article] Predicting Intermediate Data Points with Linear Interpolation in Excel and R
To make interpolation calculations easier, particularly for groups, I recently developed a new R package, interpolate().
First, let’s upload a dataset.
if(!require(readr)) install.packages("readr")
library(readr)
github="https://raw.githubusercontent.com/agronomy4future/raw_data_practice/refs/heads/main/chlorophyll_content_2024.csv"
df= data.frame(read_csv(url(github), show_col_types=FALSE))
print(head(df,5))
print(tail(df,5))
season crop reps days ch
1 2024 Sorghum 1 65 65.8
2 2024 Sorghum 2 65 63.0
3 2024 Sorghum 3 65 62.7
4 2024 Sorghum 4 65 61.4
5 2024 Sorghum 1 75 57.3
.
.
.
52 2024 Soybean 4 115 35.3
53 2024 Soybean 1 125 9.2
54 2024 Soybean 2 125 0.0
55 2024 Soybean 3 125 7.2
56 2024 Soybean 4 125 6.1This dataset contains chlorophyll content measurements for sorghum and soybean. I measured chlorophyll content every 10 days between 65 and 125 days after sowing, with four replicates at each time point.

Let’s visualize this data. First, I’ll summarize it to account for the replicates.
if(!require(dplyr)) install.packages("dplyr")
library(dplyr)
dataA= data.frame(df %>%
group_by(crop, days) %>%
dplyr::summarize(across(c(ch),
.fns= list(Mean= mean,
SD= sd,
n= length,
se= ~ sd(.)/sqrt(length(.))))))
print(dataA)
crop days ch_Mean ch_SD ch_n ch_se
1 Sorghum 65 63.225 1.851801 4 0.9259005
2 Sorghum 75 58.175 1.534872 4 0.7674362
3 Sorghum 85 56.725 2.206619 4 1.1033094
4 Sorghum 95 53.625 2.312827 4 1.1564133
5 Sorghum 105 51.300 2.493993 4 1.2469964
6 Sorghum 115 48.925 2.950000 4 1.4750000
7 Sorghum 125 46.000 4.252842 4 2.1264211
8 Soybean 65 43.675 2.991516 4 1.4957579
9 Soybean 75 42.625 2.940947 4 1.4704733
10 Soybean 85 39.450 1.190238 4 0.5951190
11 Soybean 95 37.775 1.534872 4 0.7674362
12 Soybean 105 36.100 2.102380 4 1.0511898
13 Soybean 115 30.725 4.184396 4 2.0921978
14 Soybean 125 5.625 3.963479 4 1.9817396Next, I’ll create a line graph.
if(!require(ggplot2)) install.packages("ggplot2")
library(ggplot2)
ch_contents=ggplot(data=dataA, aes(x=days, y=ch_Mean)) +
geom_errorbar(aes(ymin=ch_Mean-ch_se, ymax=ch_Mean+ch_se),
position="identity", width=3) +
geom_point(aes(fill=crop, shape=crop), size=3) +
geom_line (aes(color=crop)) +
scale_fill_manual(values=c("orange","darkred")) +
scale_shape_manual(values=c(21,21)) +
scale_color_manual(values=c("orange","darkred")) +
scale_x_continuous(breaks = seq(60, 130, 10), limits = c(60, 130)) +
scale_y_continuous(breaks = seq(0, 70, 10), limits = c(0, 70)) +
facet_wrap( ~ crop, scales="free",) +
annotate("segment", x=80, xend=110, y=Inf, yend=Inf, color="black", lwd=1) +
labs(x="Days from planting", y="Chlorophyll content (µg/cm²) ") +
theme_classic(base_size= 18, base_family = "serif") +
theme(legend.position="none",
legend.title=element_blank(),
legend.key=element_rect(color="white", fill=alpha(0.5)),
legend.text=element_text(family="serif", face="plain",
size=15, color="black"),
legend.background= element_rect(fill=alpha(0.5)),
panel.grid.major = element_line(color= "grey85", size = 0.1),
strip.background=element_rect(color="white",
linewidth=0.5,linetype="solid"),
axis.line= element_line(linewidth= 0.5, colour= "black"))
ch_contents + windows(width=9, height=5)
ggsave("C:/Users/agron/OneDrive/Desktop/Coding_Output/ch_contents.jpg",
ch_contents, width=9*2.54, height=5*2.54, units="cm", dpi=1000)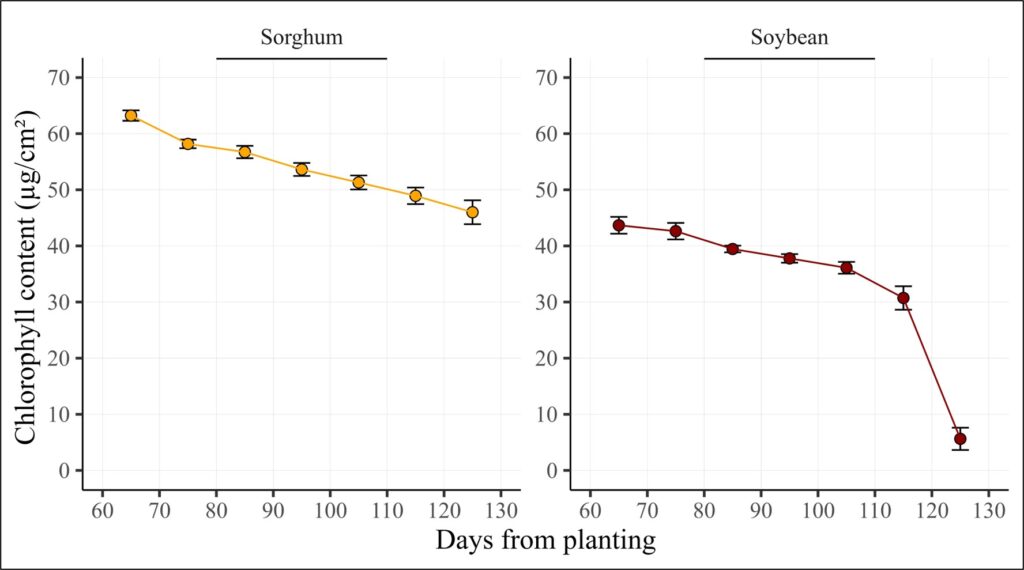
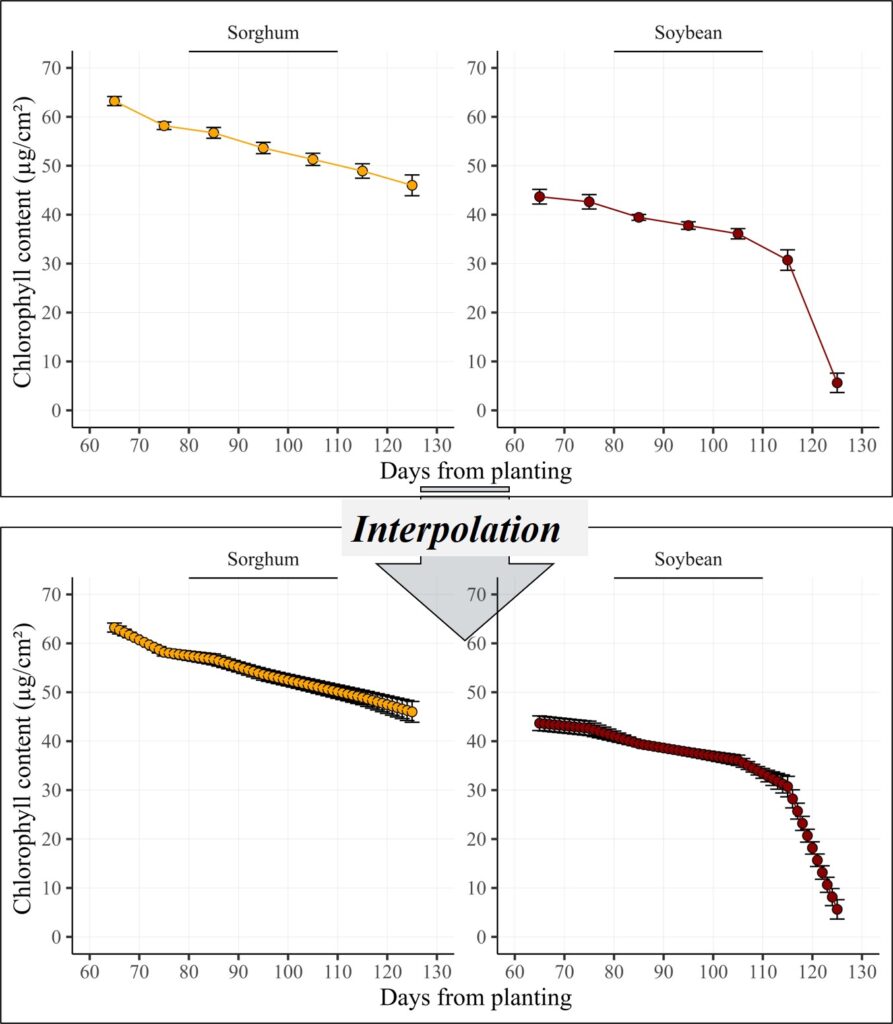
It looks good, but I want to include all data points between 65 and 125 days using interpolation. We can perform interpolation manually using Excel, R, or other programs.
■ [Data article] Predicting Intermediate Data Points with Linear Interpolation in Excel and R

However, I have developed a simple R package to make interpolation easier.
Before installing the package, please ensure that Rtools is downloaded. (https://cran.r-project.org/bin/windows/Rtools). Then, I’ll run the code below to load the package.
if(!require(remotes)) install.packages("remotes")
if (!requireNamespace("interpolate", quietly = TRUE)) {
remotes::install_github("agronomy4future/interpolate", force= TRUE)
}
library(remotes)
library(interpolate)Next, I’ll run the following code. Please specify the x and y variables for interpolation, as well as the grouping variable.
result= interpolate(df, x="days", y="ch", group_vars= c("crop","reps"))print(head(result,5))
print(tail(result,5))
crop reps season days ch category
<chr> <dbl> <dbl> <int> <dbl> <dbl>
1 Sorghum 1 2024 65 65.8 0
2 Sorghum 1 NA 66 65.0 1
3 Sorghum 1 NA 67 64.1 1
4 Sorghum 1 NA 68 63.2 1
5 Sorghum 1 NA 69 62.4 1
# A tibble: 5 × 6
crop reps season days ch category
<chr> <dbl> <dbl> <int> <dbl> <dbl>
1 Soybean 4 NA 121 17.8 1
2 Soybean 4 NA 122 14.9 1
3 Soybean 4 NA 123 11.9 1
4 Soybean 4 NA 124 9.02 1
5 Soybean 4 2024 125 6.1 0This code generates all data points between actual measurements using interpolation, grouping by crop and reps. It also creates a new column, ‘category,’ which identifies actual data points (0) and interpolated data points (1).
Next, let’s visualize this data again.
if(!require(dplyr)) install.packages("dplyr")
library(dplyr)
dataB= data.frame(result %>%
group_by(crop, days) %>%
dplyr::summarize(across(c(ch),
.fns= list(Mean= mean,
SD= sd,
n= length,
se= ~ sd(.)/sqrt(length(.))))))
print(head(dataB,5))
print(tail(dataB,5))
crop days ch_Mean ch_SD ch_n ch_se
1 Sorghum 65 63.225 1.8518009 4 0.9259005
2 Sorghum 66 62.720 1.5925242 4 0.7962621
3 Sorghum 67 62.215 1.3482211 4 0.6741105
4 Sorghum 68 61.710 1.1286570 4 0.5643285
5 Sorghum 69 61.205 0.9511221 4 0.4755611
crop days ch_Mean ch_SD ch_n ch_se
118 Soybean 121 15.665 2.553501 4 1.276750
119 Soybean 122 13.155 2.752992 4 1.376496
120 Soybean 123 10.645 3.076248 4 1.538124
121 Soybean 124 8.135 3.489035 4 1.744518
122 Soybean 125 5.625 3.963479 4 1.981740if(!require(ggplot2)) install.packages("ggplot2")
library(ggplot2)
ch_contents=ggplot(data=dataB, aes(x=days, y=ch_Mean)) +
geom_errorbar(aes(ymin=ch_Mean-ch_se, ymax=ch_Mean+ch_se),
position="identity", width=3) +
geom_point(aes(fill=crop, shape=crop), size=3) +
geom_line (aes(color=crop)) +
scale_fill_manual(values=c("orange","darkred")) +
scale_shape_manual(values=c(21,21)) +
scale_color_manual(values=c("orange","darkred")) +
scale_x_continuous(breaks = seq(60, 130, 10), limits = c(60, 130)) +
scale_y_continuous(breaks = seq(0, 70, 10), limits = c(0, 70)) +
facet_wrap( ~ crop, scales="free",) +
annotate("segment", x=80, xend=110, y=Inf, yend=Inf, color="black", lwd=1) +
labs(x="Days from planting", y="Chlorophyll content (µg/cm²) ") +
theme_classic(base_size= 18, base_family = "serif") +
theme(legend.position="none",
legend.title=element_blank(),
legend.key=element_rect(color="white", fill=alpha(0.5)),
legend.text=element_text(family="serif", face="plain",
size=15, color="black"),
legend.background= element_rect(fill=alpha(0.5)),
panel.grid.major = element_line(color= "grey85", size = 0.1),
strip.background=element_rect(color="white",
linewidth=0.5,linetype="solid"),
axis.line= element_line(linewidth= 0.5, colour= "black"))
ch_contents + windows(width=9, height=5)
ggsave("C:/Users/agron/OneDrive/Desktop/Coding_Output/ch_contents.jpg",
ch_contents, width=9*2.54, height=5*2.54, units="cm", dpi=1000)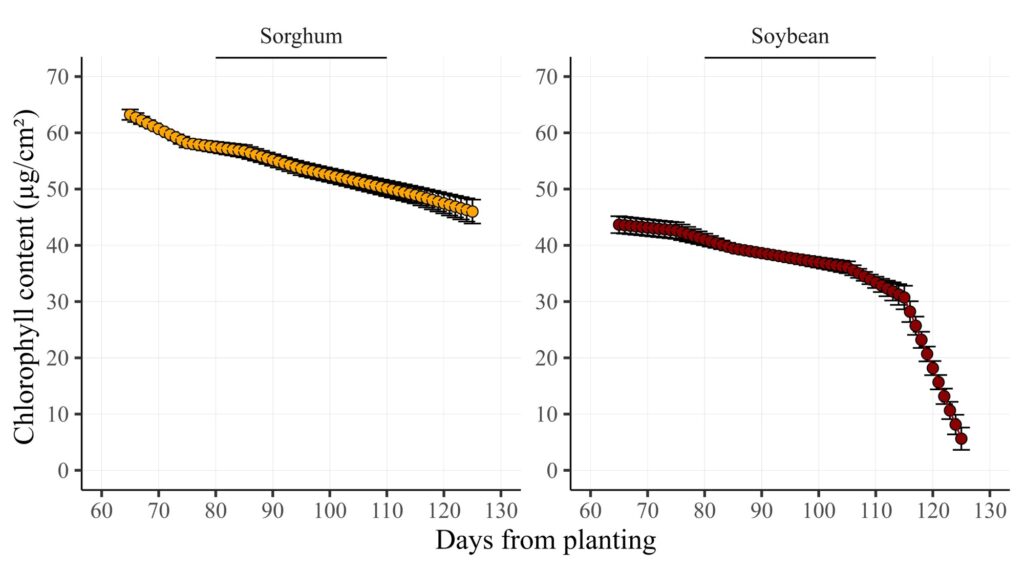
All data points have been generated between the actual measurements. This method is useful for presenting time series data, ensuring a continuous and intact representation over time.
Github: https://github.com/agronomy4future/interpolate

We aim to develop open-source code for agronomy ([email protected])
© 2022 – 2025 https://agronomy4future.com – All Rights Reserved.
Last Updated: 01/03/2025