Data filtering using R Studio
When you conduct statistical analysis, you might want to include/exclude some variables. For example, here is one data.
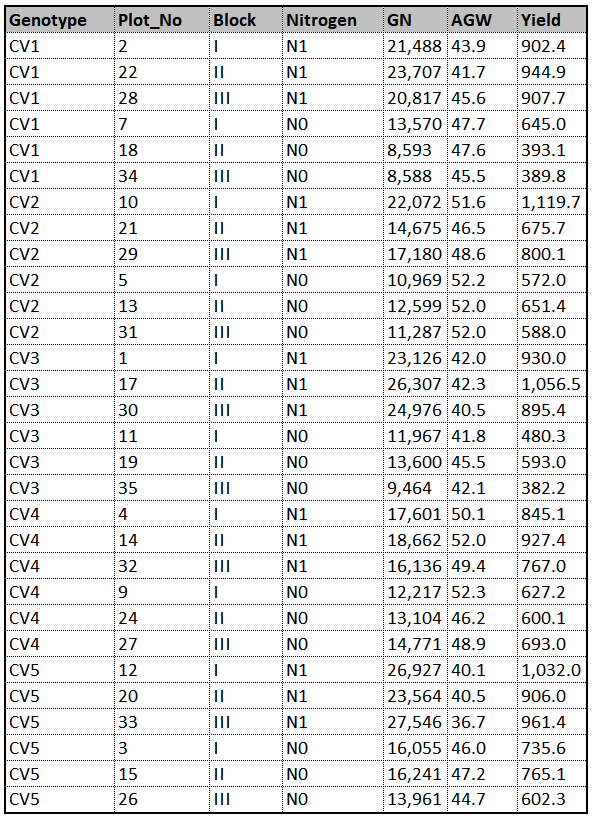
This is data about how yield, grain number (GN) and average grain weight (AGW) are different according to two different fertilizers (N0, N1) in five genotypes (CV1 – CV5). That is, there will be 10 treatments [Genotype (5) x Nitrogen (2) =10]. Replicates are 10 as blocks, and therefore experimental unit will be 30 [10 treatments x 3 blocks = 30].
What if we want to analyze in only N1 condition? or only about CV1? I’ll introduce how to filter data in R studio?
Let’s upload the data above.
# to upload data
if(!require(readr)) install.packages("readr")
library (readr)
github="https://raw.githubusercontent.com/agronomy4future/raw_data_practice/main/yield%20component_nitrogen.csv"
dataA=data.frame(read_csv(url(github),show_col_types=FALSE))
head(dataA, 3)
Genotype Block Nitrogen GN AGW Yield
1 CV1 I N1 21488 43.9 902.4
2 CV1 II N1 23707 41.7 944.9
3 CV1 III N1 20817 45.6 907.7
.
.
.1. subset
I’d like to filter one variable. For example, there are two ways to select N1.
N1= subset (dataA, Nitrogen=="N1")
N1= subset (dataA, Nitrogen!="N0")
head(N1, 5)
Genotype Block Nitrogen GN AGW Yield
1 CV1 I N1 21488 43.9 902.4
2 CV1 II N1 23707 41.7 944.9
3 CV1 III N1 20817 45.6 907.7
7 CV2 I N1 22072 51.6 1119.7
8 CV2 II N1 14675 46.5 675.7
.
.
.How about selecting several variables? For example, I want to select CV1 and N1.
CV1_N1= subset (dataA, Genotype=="CV1" & Nitrogen=="N1")
print(CV1_N1,5)
Genotype Block Nitrogen GN AGW Yield
1 CV1 I N1 21488 43.9 902.4
2 CV1 II N1 23707 41.7 944.9
3 CV1 III N1 20817 45.6 907.7How about selecting one variable and excluding another variable?
CV1X_N1= subset (dataA, Genotype!="CV1" & Nitrogen=="N1")
print(CV1X_N1)
Genotype Block Nitrogen GN AGW Yield
7 CV2 I N1 22072 51.6 1119.7
8 CV2 II N1 14675 46.5 675.7
9 CV2 III N1 17180 48.6 800.1
13 CV3 I N1 23126 42.0 930.0
14 CV3 II N1 26307 42.3 1056.5
15 CV3 III N1 24976 40.5 895.4
19 CV4 I N1 17601 50.1 845.1
20 CV4 II N1 18662 52.0 927.4
21 CV4 III N1 16136 49.4 767.0
25 CV5 I N1 26927 40.1 1032.0
26 CV5 II N1 23564 40.5 906.0
27 CV5 III N1 27546 36.7 961.4How about selecting two variables within the same factor? For example, I want to select both CV1 and CV3. So, I used the below code.
CV1_CV3= subset (dataA, Genotype=="CV1" & Genotype=="CV3")
print(CV1_CV3)
[1] Genotype Block Nitrogen GN AGW Yield
<0 rows> (or 0-length row.names)But I can’t select any variables. This is because if I select CV1, and now only CV1 exists. In this condition, if I select CV3 which does not exist, no variables are selected.
We can solve this problem using |
CV1_CV3= subset (dataA,Genotype=="CV1" | Genotype=="CV3")
print(CV1_CV3)
Genotype Block Nitrogen GN AGW Yield
1 CV1 I N1 21488 43.9 902.4
2 CV1 II N1 23707 41.7 944.9
3 CV1 III N1 20817 45.6 907.7
4 CV1 I N0 13570 47.7 645.0
5 CV1 II N0 8593 47.6 393.1
6 CV1 III N0 8588 45.5 389.8
13 CV3 I N1 23126 42.0 930.0
14 CV3 II N1 26307 42.3 1056.5
15 CV3 III N1 24976 40.5 895.4
16 CV3 I N0 11967 41.8 480.3
17 CV3 II N0 13600 45.5 593.0
18 CV3 III N0 9464 42.1 382.2Or below code is possible.
CV1_CV3= subset (dataA, Genotype!="CV2" & Genotype!="CV4" & Genotype!="CV5") How about selecting two variables within the same factor, and another variable? For example, I want to select both CV1 and CV3, and then select N1.
CV1_CV3_N1= subset (dataA, c(Genotype=="CV1" | Genotype=="CV3") & Nitrogen=="N1")
print(CV1_CV3_N1)
Genotype Block Nitrogen GN AGW Yield
1 CV1 I N1 21488 43.9 902.4
2 CV1 II N1 23707 41.7 944.9
3 CV1 III N1 20817 45.6 907.7
13 CV3 I N1 23126 42.0 930.0
14 CV3 II N1 26307 42.3 1056.5
15 CV3 III N1 24976 40.5 895.4There are no specific answers. Simply we can use below code.
N1= subset (dataA, Nitrogen=="N1")
CV1_CV3_N1= subset(N1, Genotype=="CV1" | Genotype=="CV3") First, we can select N1, and in N1, we can selecte both CV1 and CV3. We can shorten the code like below.
CV1_CV3_N1= subset(subset (dataA, Nitrogen=="N1"), Genotype=="CV1" | Genotype=="CV3")In summary, these three codes are the same code to select CV1, CV3, N1.
#1
CV1_CV3_N1= subset (dataA, c(Genotype=="CV1" | Genotype=="CV3") & Nitrogen=="N1")
#2
N1<- subset (dataA, Nitrogen=="N1")
CV1_CV3_N1= subset(N1, Genotype=="CV1" | Genotype=="CV3")
#3
CV1_CV3_N1= subset(subset(dataA, Nitrogen=="N1"), Genotype=="CV1" | Genotype=="CV3")2. dplyr package
Now, let’s use dplyr package.
if(!require(dplyr)) install.packages("dplyr")
library (dplyr)Now, I’d like to select N1
#1
dataB= dataA %>% filter (Nitrogen=="N1")
#2
dataB= dataA %>% filter (Nitrogen!="N0")
print (dataB)
Genotype Block Nitrogen GN AGW Yield
1 CV1 I N1 21488 43.9 902.4
2 CV1 II N1 23707 41.7 944.9
3 CV1 III N1 20817 45.6 907.7
4 CV2 I N1 22072 51.6 1119.7
5 CV2 II N1 14675 46.5 675.7
6 CV2 III N1 17180 48.6 800.1
7 CV3 I N1 23126 42.0 930.0
8 CV3 II N1 26307 42.3 1056.5
9 CV3 III N1 24976 40.5 895.4
10 CV4 I N1 17601 50.1 845.1
11 CV4 II N1 18662 52.0 927.4
12 CV4 III N1 16136 49.4 767.0
13 CV5 I N1 26927 40.1 1032.0
14 CV5 II N1 23564 40.5 906.0
15 CV5 III N1 27546 36.7 961.4How about selecting CV1 and N1? It’s similar with subset()
#subset()
CV1_N1= subset (dataA, Genotype=="CV1" & Nitrogen=="N1")
#dplyr
CV1_N1= dataA %>% filter (Genotype=="CV1" & Nitrogen!="N0")How about selecting both CV1 and CV3?
#subset()
CV1_CV3= subset (dataA, Genotype=="CV1" | Genotype=="CV3")
#dplyr
CV1_CV3= dataA %>% filter(Genotype=="CV1"| Genotype=="CV3")How about selecting both CV1 and CV3, and then N1?
#subset()
CV1_CV3_N1= subset (dataA, c7(Genotype=="CV1" | Genotype=="CV3") & Nitrogen=="N1")
#dplyr
CV1_CV3_N1= dataA %>% filter (c(Genotype=="CV1" | Genotype=="CV3") & Nitrogen=="N1")Now, let’s export this data to Excel.
if(!require(writexl)) install.packages("writexl")
library (writexl)
write_xlsx(CV1_CV3_N1,"C://Users/Usuari/Desktop//CV1_3_N1.xlsx")
We aim to develop open-source code for agronomy ([email protected])
© 2022 – 2025 https://agronomy4future.com – All Rights Reserved.
Last Updated: 01/03/2025
Your donation will help us create high-quality content.
PayPal @agronomy4furure / Venmo @agronomy4furure / Zelle @agronomy4furure